
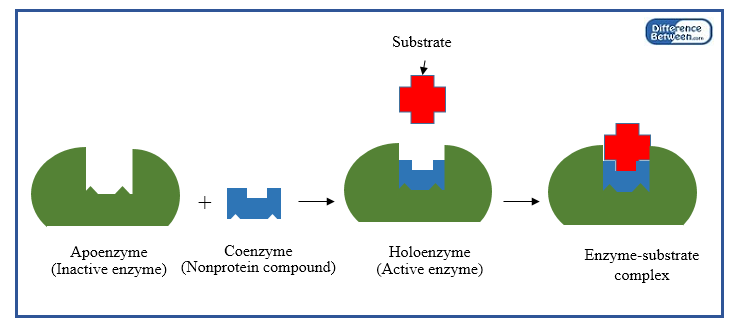
FBEB generates reduced ferredoxin, which can be oxidized to produce energy. Some of these proteins provide an alternative supply of energy to anaerobic microbes through a process known as Flavin-based electron bifurcation (FBEB) (Lubner et al., 2017). Many oxygen sensitive proteins are required for organisms to thrive in an anoxic environment. Keywords: HDX-MS, Mass spectrometry, Anaerobic proteins, H/D exchange, Protein dynamics, Protein-protein interactions, Protein-ligand interactions

Additionally, movement isn’t limited like it is in an anaerobic chamber, ensuring more consistent data, and fewer errors during the course of the reaction. To counteract this, a double vial system was developed that allows the preparation of solutions and reaction mixtures anaerobically, but also allows these solutions to be moved to an aerobic environment while shielding the solutions from oxygen. Exposure to oxygen causes the cofactors bound to these proteins, a common example being FeS clusters, to no longer interact with the amino acid residues responsible for coordinating the FeS clusters, causing loss of the clusters and irreversible inactivation of the protein. This can affect the seal, increasing the likelihood of oxygen exposure. A problem when using HDX-MS to study anaerobic proteins is that there are many parts that require constant movement into and out of an anaerobic chamber. Applying this technique to anaerobic proteins provides insight into the mechanism of proteins that perform oxygen sensitive chemistry. HDX-MS is a powerful tool for studying the protein structure-function relationship.

To further research, users can also mold the different algorithms to their needs: spectrum annotation, sequence matching, peptide variants, fraction analysis, and other such algorithms and methods can be modified to better fit your workflow.The protocol detailed here describes a way to perform hydrogen deuterium exchange coupled to mass spectrometry (HDX-MS) on oxygen sensitive proteins.
What's more, the Spectrum Maching menu allows one to input their proteomics modifications: fixed and variable modifications can be added, and you'll also have to input additional data under the Protease & Fragmentation section. Users can go very in-depth with the Enzyme alterations, too, as they can designate the cleavage rules that best meet their demands. Users are able to modify the search engines' settings: specify minimum and maximum values for peptide lengths and precursor masses, select your preferred fragmentation method, change the output format, and more. You'll first have to set up some search settings: first, input a name, then select the relevant Spectrum and Database files, as well as an output folder for exporting. Of course, a user not in tune with some knowledge related to proteomics will be a confused user: from being able to tweak the search engines and algorithms, to the managing of the Enzyme and Modifications menus, there's plenty of versatility to the program. Though a first impression might give one the feeling that the program is deceptively simple, the truth is that there's a lot of to tap into. In an effort to provide a means to streamline proteomics research, SearchGUI comes in the form of an interface capable of offering users a place where one can identify anything proteomics-related, such as peptides and proteins, through the combined work of several search engines and de novo algorithms. When conducting research on specialized topics that are harder to access by the general population, dedicated tools designed to help with such endeavors exist to proper research further, as well as provide an environment for those looking to study and deepen their knowledge on the matter.


 0 kommentar(er)
0 kommentar(er)
